Research in
the Christendat Lab
Plants produce a tremendous variety of aromatic compounds compared to other living organisms. Most of these compounds, referred to as central metabolites, are synthesized from a product of the shikimate pathway. Central metabolites are important for the structural integrity of plants and their protection from invading organisms. Other central metabolites include flavonoids and isoflavonoids, which are potential antioxidants with important nutritional benefits to humans. The regulation of the shikimate pathway is highly coordinated with the biosynthesis of these metabolites in plants. Some questions being addressed in our lab are: What are some of the biological cues that are involved in the regulation of the pathway? How can we take advantage of these regulatory processes to stimulate the shikimate pathway to enhance the biosynthesis of certain classes of central metabolites, especially those that are of nutritional importance?
The shikimate pathway is also an attractive target for drug development because it is absent in humans, but is essential for the survival of microbes, fungi and likely apicomplexan parasites. These parasites include, Plasmodium falciparum, associated with the deadliest form of malaria, and Toxoplasma gondii, implicated in psychological disorders and toxoplasmosis. We are actively studying compounds that are potential inhibitors of enzymes of the shikimate pathway with the aim of developing novel drug compounds. As a protein biochemistry group, we utilize biochemical and biophysical approaches, such as recombinant protein production, protein modification, enzyme kinetics, protein crystallography, protein ligand screening, structural biology, proteomics, etc. to understand how proteins function and how we can modulate their activity.
Ongoing Projects
Establishing Strategies to Control Agrobacterium tumefaciens Propagation in Plants
Agrobacterium tumefaciens is a gram-negative plant pathogen with the ability to infect a wide variety of hosts including, grapevines, nut trees, fruit trees, and other woody plants. This pathogen can result in severe economic losses due to its persistence and ability to infect many plant species. A. tumefaciens infection causes Crown Gall disease as plants form tumors in response to the infection. This interferes with the transportation of nutrients and phenolics within the plant. The ability for A. tumefaciens to survive within its host depends on its ability to utilize plant metabolites, especially phenylpropanoids, which are broken down through the β-ketoadipate pathway, for energy and as such, the organism is thought to hijack the host metabolome. Interestingly, a putative ATP sensing protein was identified with a gene cluster for the β-ketoadipate pathway. The biochemical mechanism via which this protein senses and regulate ATP synthesis is not well understood. Further, we determined that this protein is found in all organisms that depends on carbon uptake for ATP production. We therefore proposed that in A. tumefaciens this protein may sense low ATP levels to initiate the breakdown of aromatic compounds via the β-ketoadipate pathway which feeds acetyl CoA and succinyl CoA molecules into the tricyclic acid cycle for energy. This study will establish strategies to control A. tumefaciens infection and propagation in plants.

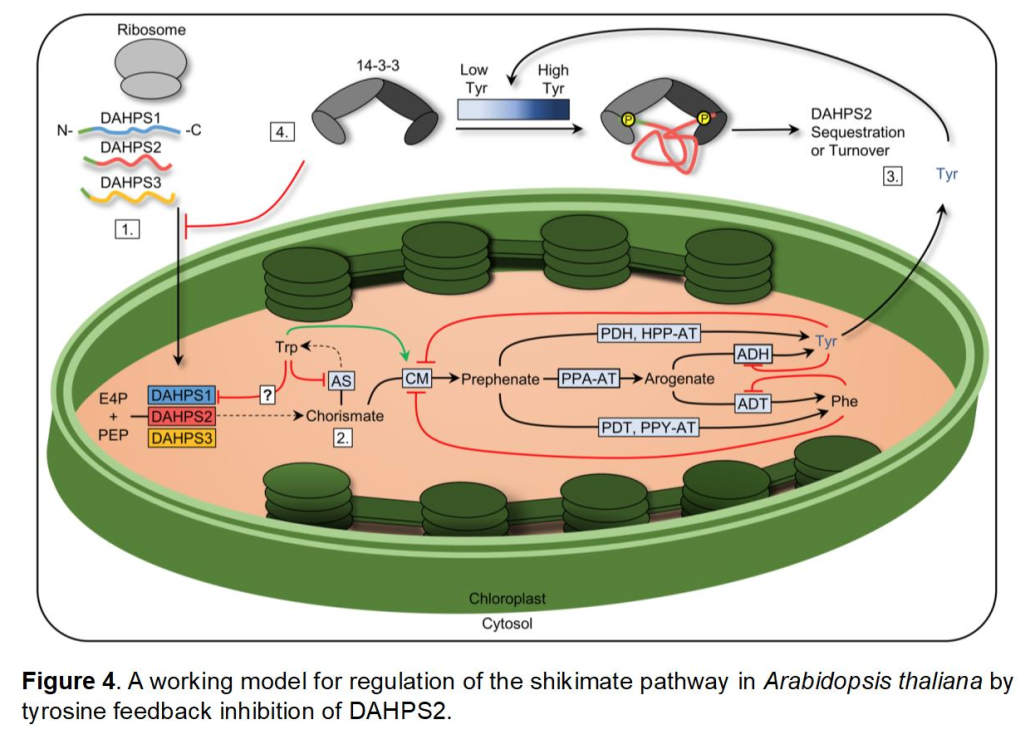
Regulation of the Shikimate Pathway Mediated by DAHPS
Entry into the shikimate pathway is mediated by 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (DAHPS). Conventional knowledge of shikimate pathway regulation involves allosteric feedback regulatory mechanisms, elucidated in a number of bacterial and fungal microorganisms. This includes allosteric feedback by the aromatic amino acids tyrosine, phenylalanine, and tryptophan, in addition to other metabolites such as chorismate and prephenate.
To date, little has been determined concerning the regulation of DAHPS within plants, therefore special interests in our lab has been the investigation of this enzyme using the model plant Arabidopsis thaliana. In Arabidopsis, three DAHPS isozymes are encoded within its genome (DAHPS1, DAHPS2, DAHPS3). Utilizing T-DNA knockout lines, we describe a hypersensitive dahps1 mutant, characterized by highly reduced root lengths caused by the aromatic amino acid tyrosine.
Supported by a number of biochemical characterizations, we established that the shikimate pathway is restricted for dahps1 seedlings in the presence of tyrosine. Interestingly, kinetic analysis did not indicate allosteric feedback as a mechanism for regulation of any of the three Arabidopsis isozymes in vitro. However, we provided initial support for a post-translational mechanism first identified in fluorescent reporter studies of DAHPS2, suggesting that this enzyme is specifically targeted for regulation in the presence of tyrosine.
We provided additional support for this mechanism based on the identification of protein-protein interactions with 14-3-3w. The regulatory role of 14-3-3 proteins are well-established in most eukaryotic organisms, especially in plants by which they act as key responders to a number of metabolites, including phytohormones (Camoni et al. 2018). The interaction between DAHPS2 and 14-3-3w is influenced by key sequence motifs that are specific for recognition by 14-3-3 proteins. Importantly, mutagenesis of identified motifs in DAHPS2 alleviates responsiveness to tyrosine.
DAHPS2 interaction with a 14-3-3 protein is a first-described case within the literature for regulation of the shikimate pathway. The modulation of DAHPS2 levels in the chloroplast by tyrosine is likely to include a number of molecular players that have yet to be identified in this regulatory network, including the possibility of kinases and phosphatases that may act upstream of the interaction of 14-3-3w with DAHPS2.
Investigating the Conservation of ATP Synthesis Across Lineages
Prokaryotes rely on the sensing and utilization of carbon compounds from their environment for ATP production. Eukaryotes, however, depend on glycolysis which couples carbon utilization with mitochondrial oxidative phosphorylation for ATP synthesis. At the molecular level, mitochondria share a common evolutionary origin with bacteria and retain features for energy sensing and ATP production. In line with this proposal, we found conserved gene clusters in bacteria that are associated with the import and utilization of carbon compounds as their energy source. Bioinformatics analysis revealed some of these genes are conserved across lineages; organisms that need to uptake carbon compounds for ATP synthesis. I am actively conducting structural biology of proteins encoded by a representative number of these conserved genes to understand how they are involved in the regulation of ATP production. In concert, we obtained the AlphaFold structure of the homologous mammalian protein and shows that it shares a highly conserved ferredoxin-like fold with the microbial protein despite sharing low sequence conservation and they adopt a similar homo-octameric oligomeric structure with 8-equivalent ATP binding pockets. These observations are congruent with the proposed role of these proteins as energy sensors and modulators of ATP production.

Investigating the Evolution of Function of SKL1 in Chloroplast Biogenesis
The shikimate pathway is a fundamental component of aromatic amino acid biosynthesis in plants and microorganisms. The seven enzymatic steps of the pathway redirect the flow of organic carbon from primary metabolism towards the production of chorismate. The functional diversification of the fifth enzyme in this pathway, shikimate kinase (SK), represents a unique expansion of molecular complexity in terrestrial plants. Prior works from our lab show that shikimate kinase-like 1 (SKL1) is required for chloroplast biogenesis and is potentially involved in state transition. The Arabidopsis thaliana SKL1-deficient mutation, skl1-8, exhibits an albino phenotype, consistent with mutational defects affecting chloroplast assembly. Chemical complementation of this mutation with the ancient homologue Marchantia polymorpha SKL1 reveals perturbations in thylakoid membranes that are consistent with shade-adapted chloroplasts. Although compelling evidence exists that SKL1 performs an evolutionarily novel role from SK, its precise molecular function remains undetermined. We are investigating the function of SKL1 to understand it evolutionary trajectory in chloroplast biogenesis.

Protocatechuate Biosynthesis in L. monocytogenes
The protocatechuate pathway branches away from the shikimate pathway and provides a route for microorganisms to metabolize plant biomass. Though most microorganisms catabolize protocatechuate as a carbon energy source, the foodborne pathogen, Listeria monocytogenes, converts shikimate and quinate to protocatechuate but is unable to utilize it. L. monocytogenes protocatechuate metabolism is controlled by a distinct set of shikimate pathway associated genes located within the qui1 and qui2 operons. Our lab seeks to understand the biological role of this pathway by investigating genes from the qui1 and qui2 operons concomitantly with the associated bacterial regulators, QuiR and QuiR2. We uncover novel bacterial metabolic pathways that produce aromatic compounds that are important for microbial communications. To accomplish this, we are further exploring these compounds to better characterize microbial interactions.

Alternation in Microbial Communities of Listeria monocytogenes through Biofilm
Listeria monocytogenes is a prominent soil-borne pathogen known globally for its ability to survival in extreme environments and its potential to cause human disease, listeriosis. A significant concern is its ability to form biofilms on food and processing surfaces, making surface decontamination challenging which increases the risk of frequent outbreaks. Previous studies showed listeria monocytogenes persistence is facilitated by its ability to co-exist with other organisms, such as pseudomonas putida. The shikimate pathway is known for its role in the biosynthesis of aromatic compounds in microorganisms and plants and is involved in biofilm formation. On the other hand, it was shown that excess shikimate inhibits biofilm formation in some soil-borne bacteria, including Stenotrophomonas maltophilia (Isom et al., 2021). These findings, indicating an association between shikimate pathway and biofilm formation, led us to hypothesize that shikimate would influence microbial interaction in the environment. My interest lies in the role of shikimate on biofilm formation in microbiome conditions starting with co-culture using Listeria and Pseudomonas species. It is expected that findings from these studies will provide innovative approaches to decontaminate food processing surfaces to reduce listeria outbreak incidents in agriculture and the food industry.
